Authors: Ken Nakae1, Misako Komatsu2*, Takaaki Kaneko3,4,5, Junichi Hata3,4,5, Hideyuki Okano3,4, Shin Ishii1, Tetsuo Yamamori2
*corresponding author: Misako Komatsu (mskkomatsu@gmail.com)
DATASET DESCRIPTION
This page provides annotated electrode layouts for the animals that participated in ECoG experiments.
Method
The positions of electrodes of the electrocorticography (ECoG) for marmosets are normalized to the standard space, which is defined using the standard brain, BMA 2019 Ex vivo, in Brain/MINDS data portal.
This normalization consists of two steps: 1. electrode identification on the individual in vivo MRI (subject space) and 2. registration to the averaged ex vivo MRI (standard space). Here, we explain these two steps.
1. Electrode identification on the subject space
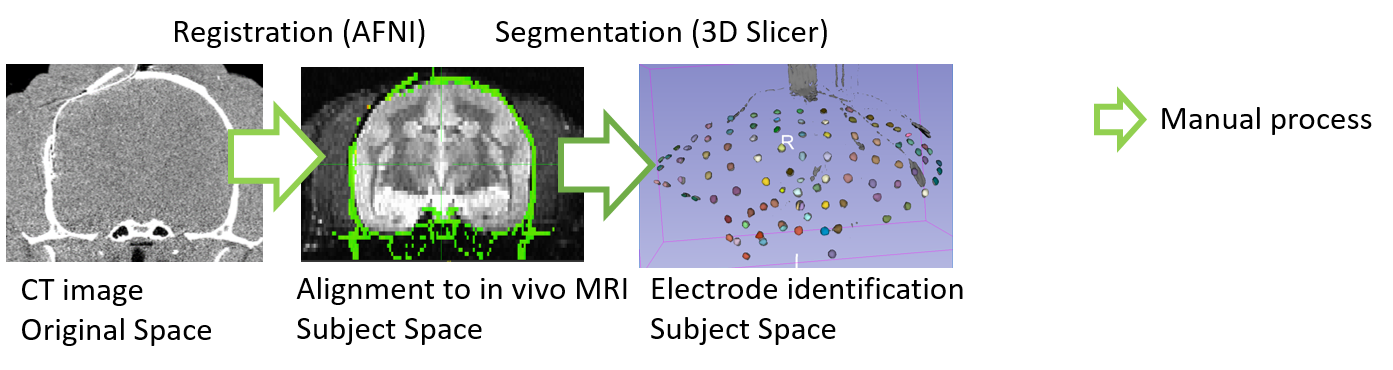
We obtain a computed tomography (CT) scan image of individual marmoset after an electrode implantation, and an in vivo T2 weighted magnetic resonance imaging (MRI) for the same individual marmoset before surgery. The CT image is manually aligned to the in vivo MRI space by rigid transformation using AFNI software (Cox, 1996). The electrodes are manually segmented from the CT images using 3D Slicer (Fedorov et al., 2012) to eliminate non-electrode regions and numbering the electrodes.
2. Registration to the standard space
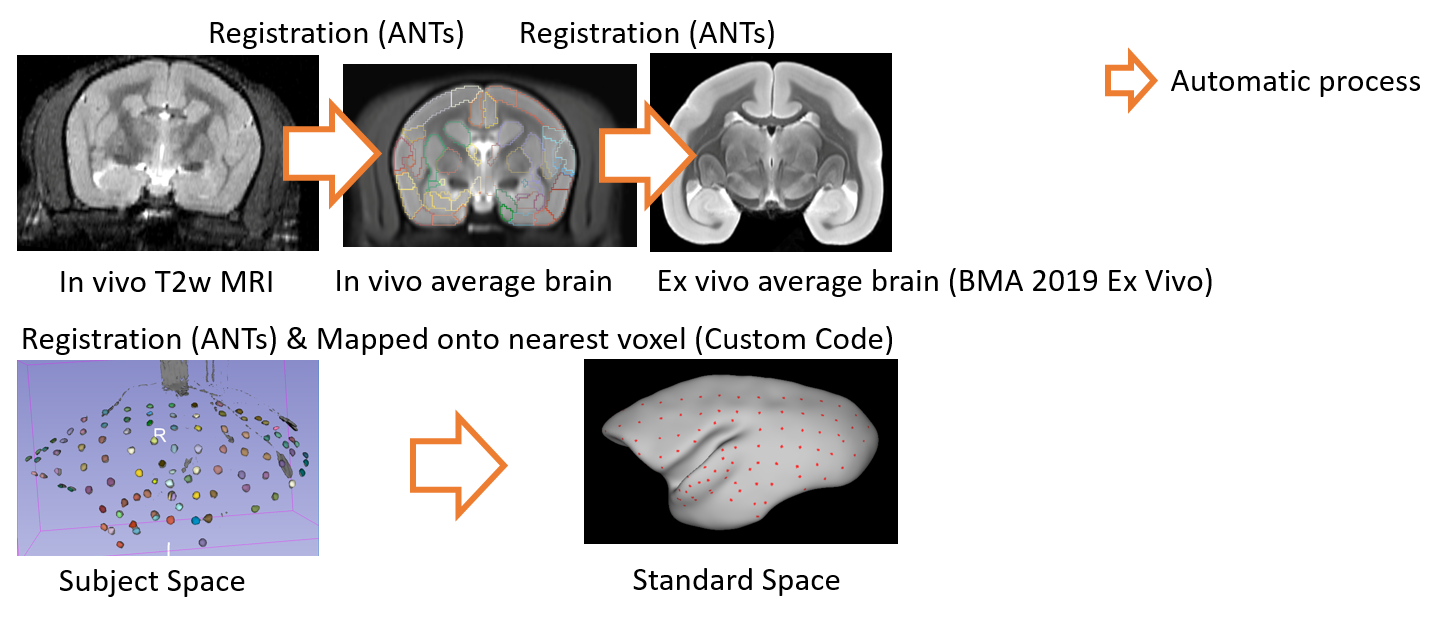
The in vivo individual MRI (subject space) is registered to the in vivo average MRI data of marmosets by free-form deformation using ANTs software (Avants et al, 2011). The in vivo averaged MRI data is also registered to the standard space defined by BMA 2019 Ex vivo using ANTs software. Then, we map the segmented electrode on the subject space to those on the standard space based on these registrations. Finally we determine a nearest voxel of each electrode on the standard brain using a nearest neighbor method, and annotate a cortical region of the voxel to the electrode.
Contents of Data
ElectrodesXx.zip contains “ElectrodesXx.mat” and “ElectrodeIndexXx.csv”.
1. ElectrodesXx.mat: Information of the ECoG electrode locations on a 2D subject space
MRI: 2D view of the marmoset brain
LINE: The outline of the marmoset brain
X: X positions of the electrodes
Y: Y positions of the electrodes
(If there are)
LX: X positions of the LEDs
LY: Y positions of the LEDs
IX: X positions of the virus injections
IY: Y positions of the virus injections
2. ElectrodesIndexXx.csv: Information of the ECoG electrode on the standard space
Column 1: segment number
Column 2: segment name
Column 3: area number based on Hashikawa Atlas
Column 4: area abbreviation
Column 5: area name
Column 6: hemisphere
Column 7-9: RAS coordinates on the standard space
CITATION
Nakae, Ken; Komatsu, Misako; Kaneko, Takaaki; Hata, Junichi; Okano, Hideyuki; Ishii, Shin; Yamamori, Tetsuo : Brain/MINDS Marmoset Brain ECoG Annotations of Electrodes (DataID: 4958)
LICENSE
 This work is licensed under a Creative Commons Attribution 4.0 International License.
This work is licensed under a Creative Commons Attribution 4.0 International License.
DOWNLOAD
2024/07/08: The data (Monkey Ca, Rm, Yo) were uploaded.
2021/09/21: The page was released.
