Brain/MINDS Marmoset Connectivity Resource
Introduction
The BMCR is a new resource that provides access to anterograde and retrograde neuronal tracer data in the prefrontal cortex of a marmoset brain. It comprises post-processed data and tools for exploring and reviewing the data, and for integration with other marmoset brain image databases and cross-species comparisons. We visualize data of different individuals in a common reference space at an unprecedented high resolution. Major features are:
- New image data of anterograde and retrograde neuronal tracers.
- dMRI tractography data that is associated with anterograde injection sites.
- Transformation fields to between other marmoset brain image databases and atlas (Brain/MINDS Marmoset Reference Atlas, Marmoset Brain Mapping, Marmoset Brain Connectivity Atlas).
- Transformation fields to the human MNI space
- Flatmap stacks of neural tracer data (Including the all neccesary data to map other image data to flatmap stacks).
- The BMCR-Explorer and the Nora-StackApp for reviewing the data
- A left-right symmetric population-average brain image template based on serial two-photon tomography along with a left-right symmetric population-average HARDI, Nissl, and backlight image.
- We integrated retrograde neural tracer data from the Marmoset Brain Connectivity Atlas project.

More information can be found on our web-page (http://bmca.riken.jp/) and in our paper:
The Brain/MINDS Marmoset Connectivity Resource: An open-access platform for cellular-level tracing and tractography in the primate brain
H. Skibbe, M.F. Rachmadi, K. Nakae, C. E. Gutierrez, J. Hata, H. Tsukada, C. Poon, K. Doya, P. Majka, M. G. P. Rosa, M. Schlachter, H. Okano, T. Yamamori, S. Ishii, M. Reisert, A. Watakabe. In PLoS Biology, 2023
Updates:
- 2024/10 : Moved the BMCR Explorer related content to a new web-page (https://bmcr.brainminds.jp/)
- 2023/06/30:
- We revised our data repository. We’ve restructured the folders to improve usability and recompressed some of the files to decrease download size. We also refined the meta data information (In particular marmoset related information such as age, sex and body weight and file descriptions). Fixed incorrect references to the MBM atlas.
- 2023/05/14:
- Added further information. Updated links. All data is now available without restrictions (Removed passwords). BMCA -> BMCR. Added license information.
- 2023/03/27:
- We fixed a bug in the Nora-stack app for MacOS where the pre-defined workspaces did not work
- We added a tutorial explaining how to download and install the Nora-StackApp
BMCR-Explorer
The BMCR-Explorer (click here) is publicly available online. An overview about its functionality is shown in the video on the right. We prepared further videos that can help you to get started (click here). You can also directly try it out by clicking on one of the three example images below.
The BMCR-Explorer works best with google-chrome (and chromium browser), but should also work with the chromium, firefox and edge browsers. Safari might work. It also has touch support. It won’t work with the Microsoft Internet Explorer :-(.
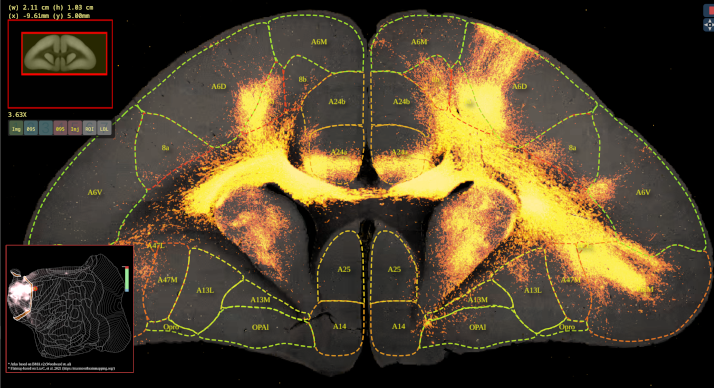
BMCR-Explorer Example 1: An anterograde tracer segmentation mask on top of the original serial two-photon tomography image (click on the figure to open the image in the BMCA-Explorer).
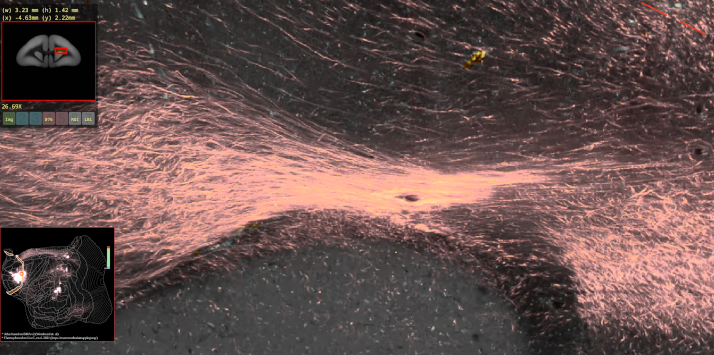
BMCR-Explorer Example 3: A serial two-photon tomography image of an anterograde neural tracer in the marmoset brain (click on the figure to open the image in the BMCA-Explorer).
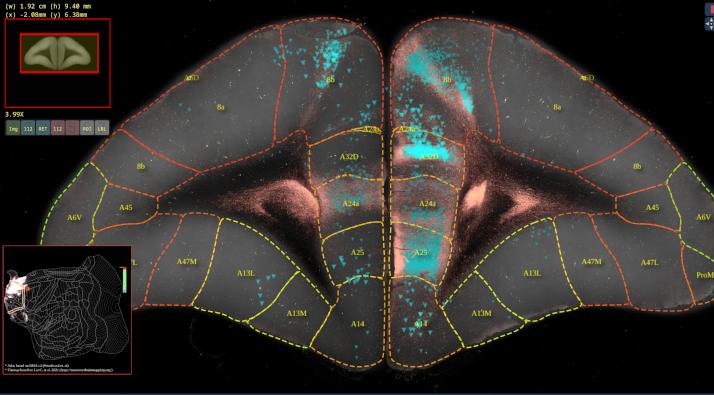
BMCR-Explorer Example 2: An anterograde and retrograde tracer (click on the figure to open the image in the BMCA-Explorer).
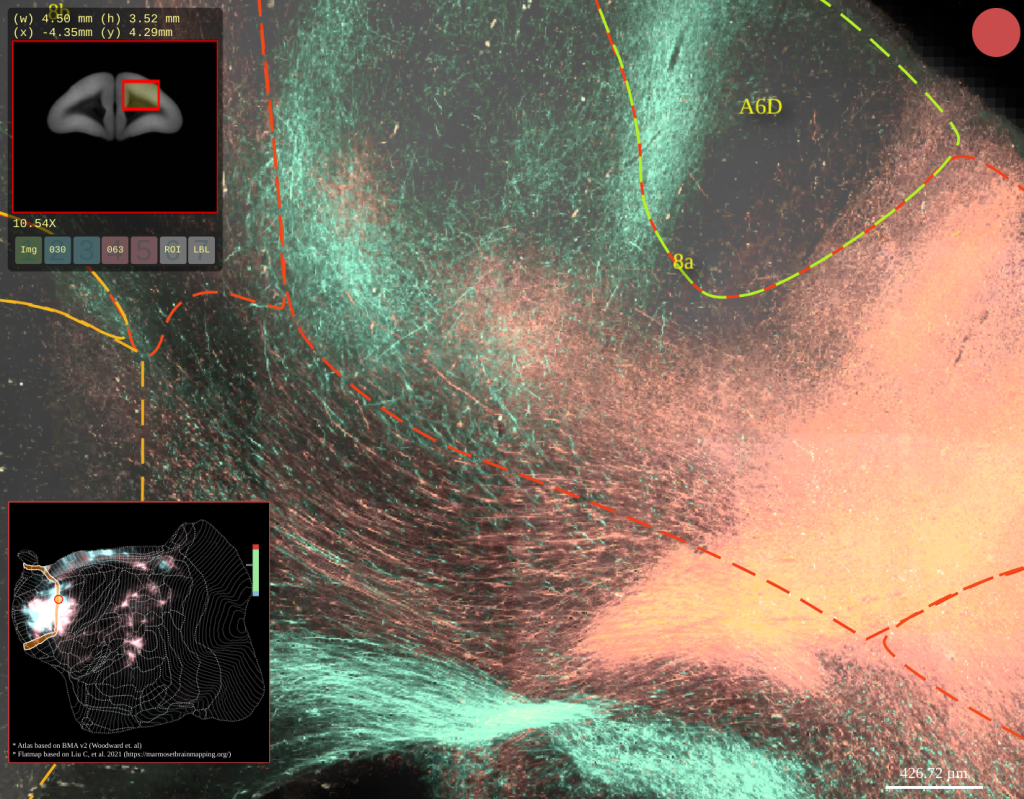
BMCR-Explorer Example 4: Two serial two-photon tomography images of anterograde neural tracers from two different marmoset brains (click on the figure to open the image in the BMCA-Explorer).
Mapping between the marmoset and the human brain
The BMCR provides a warping field between the STPT image space and the human MNI space. The video on the right shows an anterograde tracer segmentation that has been mapped from a marmoset brain to a human brain. The warping fields are publicly available (see Data-Download)
Nora-StackApp
The Nora-StackApp is based on the nora medical imaging platform, which we extended with functions so that it interacts with BMCR-Explorer and flatmap stacks. You download the app from here ->here<-, and can find instruction on how to install the Nora-StackApp here: Nora-StackApp Howto.
Flatmap-stacks mapping
A flatmap extents cortical flatmaps with cortical depth. It allows the exploration of tracer data in a coordinate system, where the plane defines the location on the marmoset cortex, and z the cortical depth. The mapping is publicly available: https://github.com/BrainImageAnalysis/flatmap_stack
Data-download & License
You can download the data from ->here<-. The dataset includes the STPT template images (with a population average STPT background image, a population average Nissl image, a population average backlit image and a population average HARDI. It further incooperates measurements derived from a population average HARDI. It also includes the ANTs warping field that maps data from our STPT template space to a flatmap-stack, and ANTs warping fields to map between other marmoset brain atlases (MARMOSET BRAIN CONNECTIVITY ATLAS, MARMOSET BRAIN MAPPING version 2 and 3, Brain/MINDS Marmoset Reference Atlas v1.1.0). It also includes the warping field between the STPT image space and the human MNI space, and a copy of the Nora-StackApp.
License:
Our image data is published under the CC BY 4.0 license (https://creativecommons.org/licenses/by/4.0/deed.en).
The downloadable dataset also includes data derived from four atlases that are work of other groups, which you might find helpful in reproducing and exploring our findings. Please carefully read the accompanying ‘Readme’ files, as some of the data is released under different licenses. If you find these data useful and plan to use it, then you must cite the original work, and accept their licenses. A summary of the atlas licenses is provided below:
- (1) Brain/MINDS 3D Marmoset Reference Brain Atlas 2019 (abbreviated as BMA)
- web-link: https://dataportal.brainminds.jp/atlas-package-download-main-page/bma-2019-ex-vivo.
- Reference:
- Woodward, Alexander; Gong, Rui; Nakae, Ken; Hata, Junichi; Okano, Hideyuki; Ishii, Shin; Yamaguchi, Yoko : Brain/MINDS 3D Marmoset Reference Brain Atlas 2019 (DataID: 4520) https://doi.org/10.24475/bma.4520
- License: Released under a Creative Commons Attribution 4.0 International License; check the webpage for details.
- (2) Data from the Marmoset Brain Connectivity Atlas (abbreviated as MBCA)
- web-link: https://www.marmosetbrain.org/reference
- References:
- The Marmoset Brain in Stereotaxic Coordinates” by Paxinos, Watson, Petrides, Rosa and Tokuno (Academic Press, 2012)
- Majka P., Bai S., Bakola S., Bednarek S., Chan J.C., Jermakow N., Passarelli L., Reser D.H., Theodoni P., Worthy K.H., Wang X-J., Wójcik D.K., Mitra P.P. & Rosa M.G.P. (2020). Open access resource for cellular-resolution analyses of corticocortical connectivity in the marmoset monkey. Nature Communications. http://doi.org/10.1038/s41467-020-14858-0
- Majka P., Chaplin T.A., Yu, H.-H., Tolpygo A., Mitra P.P., Wójcik D.K., & Rosa M.G.P. (2016). Towards a comprehensive atlas of cortical connections in a primate brain: Mapping tracer injection studies of the common marmoset into a reference digital template. Journal of Comparative Neurology, 524(11), 2161–2181. http://doi.org/10.1002/cne.24023
- Lincense: Released under a Creative Commons Attribution-ShareAlike 4.0 (CC-BY-SA) License; check the webpage for details.
- (3) Atlas data from the marmoset brain mapping project (version 2+3) (abbreviated as MBM)
- web-link: https://marmosetbrainmapping.org/atlas.html
- References:
- version 3: Liu C, et al. Marmoset Brain Mapping V3: Population multi-modal standard volumetric and surface-based templates. Neuroimage (2021) doi:10.1016/j.neuroimage.2020.117620.
- version 2: Liu C, et al. A resource for detailed 3D mapping of white matter pathways in the marmoset brain. Nature Neuroscience (2020) doi:10.1038/s41593-019-0575-0.
- Lincense: Released under a Creative Commons Attribution-NonCommercial-ShareAlike (CC BY-NC-SA 4.0) License; check the webpage for details.
Related publications:
- Local and long-distance organization of prefrontal cortex circuits in the marmoset brainA. Watakabe, H. Skibbe, K. Nakae, H. Abe, N. Ichinohe, M. F. Rachmadi, J. Wang, M. Takaji, H. Mizukami, A. Woodward, R. Gong, J. Hata, D. C. Van Essen, H. Okano, S. Ishii, T. Yamamori. In Neuron, 2023
- The Brain/MINDS Marmoset Connectivity Resource: An open-access platform for cellular-level tracing and tractography in the primate brain
H. Skibbe, M.F. Rachmadi, K. Nakae, C. E. Gutierrez, J. Hata, H. Tsukada, C. Poon, K. Doya, P. Majka, M. G. P. Rosa, M. Schlachter, H. Okano, T. Yamamori, S. Ishii, M. Reisert, A. Watakabe. In PLoS Biology, 2023 - The Brain/MINDS Marmoset Connectivity Atlas: Exploring bidirectional tracing and tractography in the same stereotaxic space
H. Skibbe, M.F. Rachmadi, K. Nakae, C. E. Gutierrez, J. Hata, H. Tsukada, C. Poon, K. Doya, P. Majka, M. G. P. Rosa, H. Okano, T. Yamamori, S. Ishii, M. Reisert, A. Watakabe. In bioRxiv, Cold Spring Harbor Laboratory, 2022. - Connectional architecture of the prefrontal cortex in the marmoset brain. Watakabe., A, Skibbe, H., Nakae, K., Abe, H., Ichinohe, N., Wang, J., Takaji, M., Mizukami, H., Woodward, A., Gong, R., Hata, J., Okano, H., Ishii, S. & Yamamori., T. bioRxiv 2021.12.26.474213; doi: https://doi.org/10.1101/2021.12.26.474213
Contact: The content of this page is maintained by the Brain Image Analysis Unit, Riken CBS. Please feel free to contact us with any questions or comments about the content. Contact information can be found on our homepage: https://bia.riken.jp/.