The Brain/MINDS 3D Digital Marmoset Brain Atlas Version 2.0
Authors: Rui Gong*1,2, Noritaka Ichinohe3, Hiroshi Abe4, Toshiki Tani4, Mengkuan Lin5, Takuto Okuno6, Ken Nakae7, Junichi Hata6,8,9, Shin Ishii2, Patrice Delmas10, Shahrokh Heidari10, Jiaxuan Wang10, Tetsuo Yamamori4, Hideyuki Okano8,9, and Alexander Woodward1
1. Connectome Analysis Unit, RIKEN Center for Brain Science, Wako, Saitama, Japan
2. Integrated Systems Biology Laboratory, Department of Systems Science, Graduate School of Informatics, Kyoto University, Kyoto, Japan
3. Department of Ultrastructural Research, National Center of Neurology and Psychiatry, Kodaira, Tokyo, Japan
4. Laboratory for Molecular Analysis of Higher Brain Function, RIKEN Center for Brain Science, Wako, Saitama, Japan
5. Department of Neurobiology, University of Pittsburgh School of Medicine, Pittsburgh, PA, United States
6. Graduate School of Human Health Sciences, Tokyo Metropolitan University, Tokyo, Japan
7. Graduate School of Engineering, University of Fukui, Fukui, Japan
8. Laboratory for Marmoset Models of Brain Diseases, RIKEN Center for Brain Science, Wako, Saitama, Japan
9. Keio University Regenerative Medicine Research Center, Kawasaki, Kanagawa, Japan
10. Intelligent Vision Systems Laboratory, The University of Auckland, Auckland, New Zealand
*corresponding author: Rui Gong (rui.gong@a.riken.jp)
DATASET DESCRIPTION
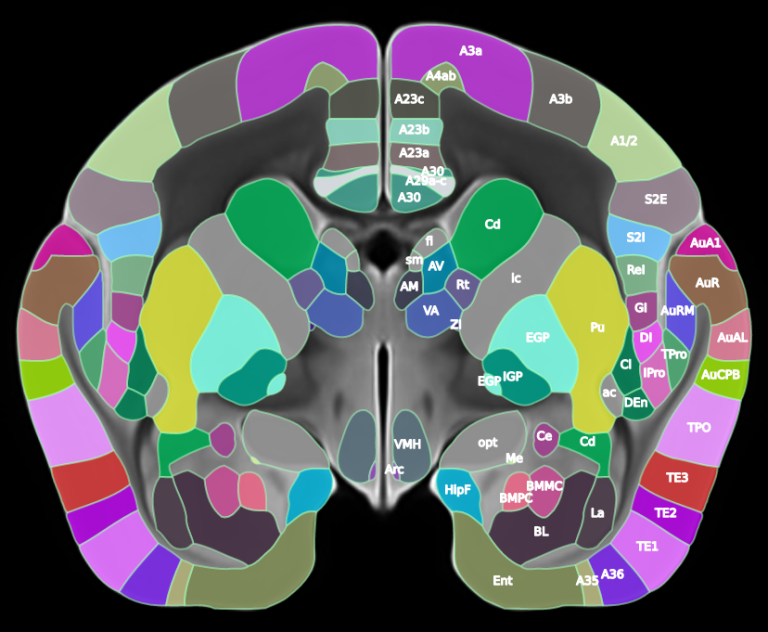
We present our new Brain/MINDS 3D digital marmoset brain atlas version 2.0 (BMA2.0), a population-based 3D digital brain atlas of the common marmoset (Callithrix jacchus), designed to overcome the limitations of previous single subject atlases that are prone to structural biases arising from individual variation. Here, manually delineated cortical regions from 10 myelin-stained brains were used to create a generalized cortical parcellation. Newly refined subcortical regions from a previous atlas and a completely new cerebellum parcellation were also incorporated, resulting in a comprehensive whole brain parcellation for both hemispheres. To facilitate multimodal data analysis, the atlas package includes co-registered average templates for myelin and Nissl staining from the same individuals, ex vivo MRI T2 (91 individuals), and in vivo MRI T2 (446 individuals). Cortical flat maps and pial, cortical mid-thickness, and white matter surfaces are also provided. BMA2.0 provides a central brain space for multimodal data integration, spatial analysis, and comparative neuroscience. Standard formats and transformations are provided for easy integration into existing workflows and interoperability with existing atlases.
LICENSE
 This work is licensed under a Creative Commons Attribution 4.0 International License.
This work is licensed under a Creative Commons Attribution 4.0 International License.
DOWNLOAD
VERSION HISTORY
- 28-09-2025: Initial release.
